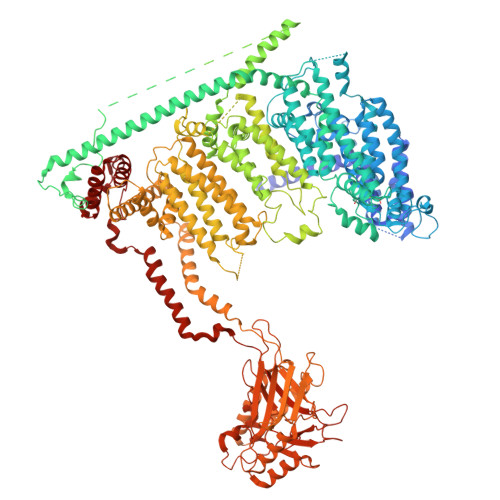
Structures of invertebrate PEZO-1 isoforms with a compact architecture and a dispensable pore-distal N-terminal blade.
Bell, B., Jaramillo-Granada, A.M., Orlin, D.J., Weng, W.H., Wen, H., Sotomayor, M., Chesler, A.T., Baker, M.L., Cordero-Morales, J.F., Vasquez, V.(2025) Cell Rep 45: 116878-116878
- PubMed: 41477764
- DOI: https://doi.org/10.1016/j.celrep.2025.116878
- Primary Citation of Related Structures:
9ZIS, 9ZIT - PubMed Abstract:
PIEZO channels are mechanosensitive ion channels conserved from plants to humans, yet structures exist for only a few mammalian orthologs. We define the structural and functional diversity of Caenorhabditis elegans PEZO-1, a single gene with extensive alternative splicing, by determining cryo-electron microscopy structures of three representative isoforms: G (full length), K (lacking the pore-distal N-terminal blade), and L (missing most of the blade). PEZO-1G displays mechanically evoked currents yet adopts a compact, semi-flattened conformation that significantly differs from the mammalian domes. The blades exhibit a three-step slope architecture stabilized by inter-blade latching among transmembrane helical units, yielding a circular, steering-wheel-like arrangement. A wider cap enables distinct blade-cap contacts that stabilize a "toggle-down" conformation. Isoform K also exhibits mechanically evoked currents, indicating that the pore-distal N-terminal blade is dispensable for mechanoactivation. Computational membrane-deformation modeling indicates that the isoforms impose distinct curvatures on the bilayer. Our findings indicate an evolutionarily distinct architecture for PEZO-1.
- Department of Biochemistry and Molecular Biology, Center for Membrane Biology, McGovern Medical School at the University of Texas Health Science Center at Houston, Houston, TX 77030, USA.
Organizational Affiliation: