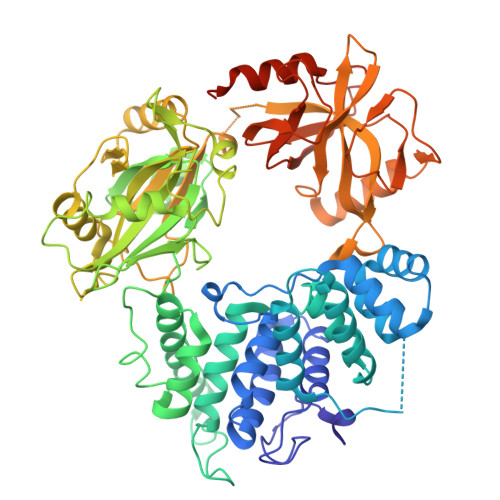
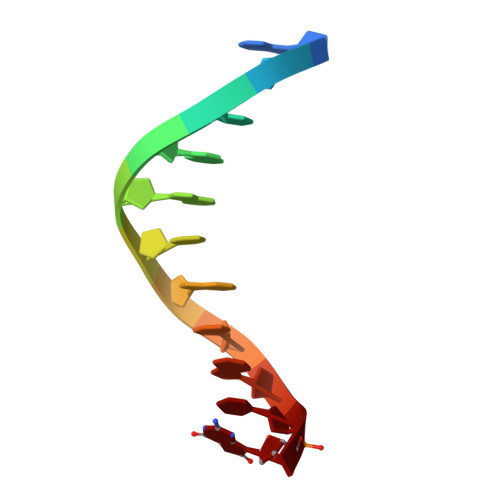
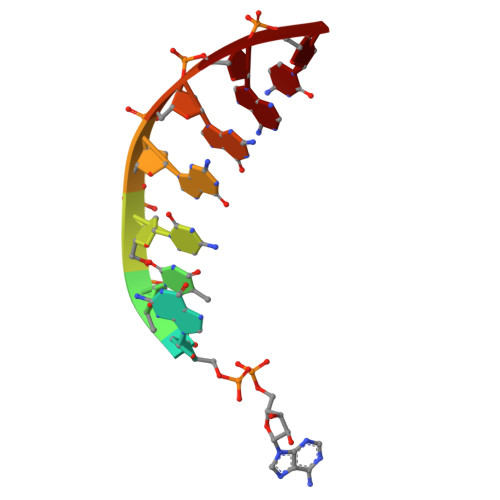
Processing of DNA single-strand breaks with oxidatively damaged ends by LIG1.
Balu, K.E., Almohdar, D., Lerner, C., Ratcliffe, J., Tang, Q., Parwal, T., Lee, K.M., Prakash, A., Caglayan, M.(2025) Nucleic Acids Res 53
- PubMed: 41370201
- DOI: https://doi.org/10.1093/nar/gkaf1344
- Primary Citation of Related Structures:
9YHU, 9YHV, 9YHW, 9YHX, 9YHY - PubMed Abstract:
DNA ligase 1 (LIG1) seals broken strand breaks by joining two adjacent ends during DNA replication and repair transactions. We previously reported atomic-level insight into the strategies that LIG1 uses to discriminate mismatches or ribonucleotides. However, how LIG1 processes strand breaks with oxidatively damaged ends in the absence and presence of a "wrong" sugar remains unknown. Here, we determined the crystal structures of LIG1/nick DNA complexes with 3'-8-oxodG and 3'-8-oxorG templating A or C during the pre- and post-catalytic steps of the ligation reaction. Our structures demonstrated differences in the distances at the +1 and +2 nucleotides relative to the 3'-end of the nick and a shift in the template base position to accommodate the oxidative lesion depending on the dual coding potential of 8-oxoG, which forms Hoogsteen or Watson-Crick base pairing in -syn or -anti conformation. Furthermore, these structural adjustments lead to mutagenic ligation or non-mutagenic end joining of the nick substrates. Overall, our findings provide mechanistic insight into how LIG1 processes nicks harboring oxidative damage and ribonucleotides to ensure fidelity at the final ligation step of DNA repair and replication to maintain genome integrity.
- Department of Biochemistry and Molecular Biology, University of Florida, Gainesville, FL 32610, United States.
Organizational Affiliation: