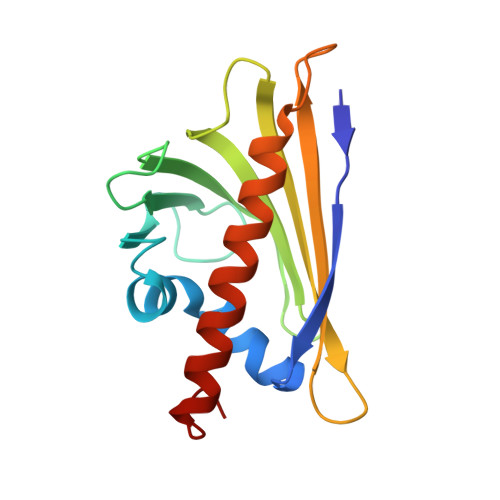
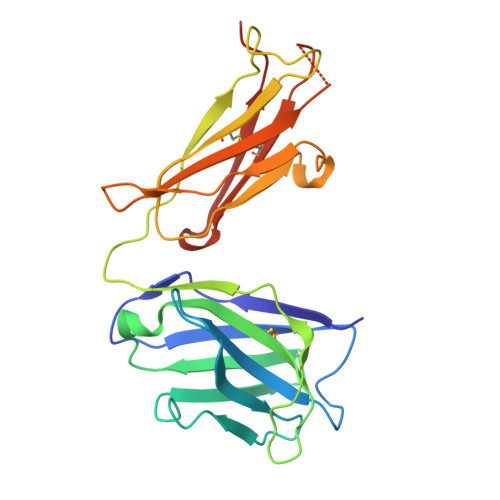
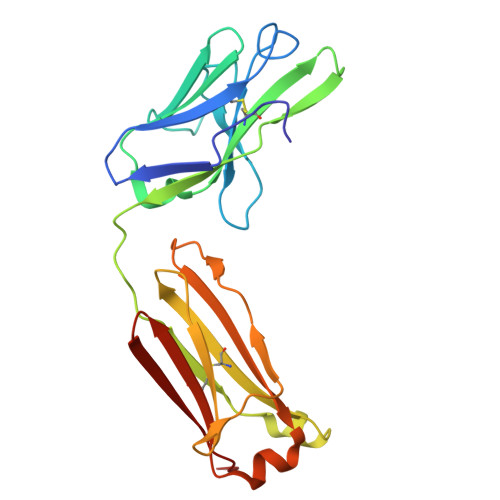
Structural Determination of a Human IgE Epitope on Major Birch Allergen Bet v 1.
O'Malley, A., Borders, B.T., Wilson, J.M., Smith, S.A., Chruszcz, M.(2026) Allergy
- PubMed: 41622586
- DOI: https://doi.org/10.1111/all.70240
- Primary Citation of Related Structures:
9Y0A, 9Y0D, 9Y0E - Department of Biochemistry and Molecular Biology, Michigan State University, East Lansing, Michigan, USA.
Organizational Affiliation: