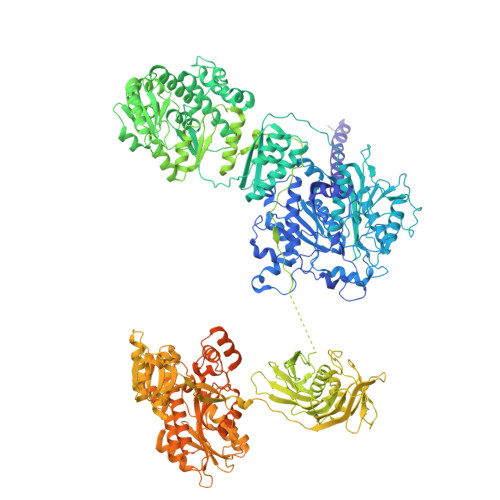
Molecular Basis for Asynchronous Chain Elongation During Rifamycin Antibiotic Biosynthesis.
Liu, C., West, R.C., Chen, M., Cohn, W., Wang, G., Mandot, A.M., Kim, S., Cogan, D.P.(2025) bioRxiv
- PubMed: 40631255
- DOI: https://doi.org/10.1101/2025.07.05.663307
- Primary Citation of Related Structures:
9PAT, 9PAV, 9PC6 - PubMed Abstract:
The rifamycin synthetase (RIFS) from the bacterium Amycolatopsis mediterranei is a large (3.5 MDa) multienzyme system that catalyzes over 40 chemical reactions to generate a complex precursor to the antibiotic rifamycin B. It is considered a hybrid enzymatic assembly line and consists of an N-terminal nonribosomal peptide synthetase loading module followed by a decamodular polyketide synthase (PKS). While the biosynthetic functions are known for each enzymatic domain of RIFS, structural and biochemical analyses of this system from purified components are relatively scarce. Here, we examine the biosynthetic mechanism of RIFS through complementary crosslinking, kinetic, and structural analyses of its first PKS module (M1). Thiol-selective crosslinking of M1 provided a plausible molecular basis for previously observed conformational asymmetry with respect to ketosynthase (KS)-substrate carrier protein (CP) interactions during polyketide chain elongation. Our data suggest that C-terminal dimeric interfaces-which are ubiquitous in bacterial PKSs-force their adjacent CP domains to co-migrate between two equivalent KS active site chambers. Cryogenic electron microscopy analysis of M1 further supported this observation while uncovering its unique architecture. Single-turnover kinetic analysis of M1 indicated that although removal of C-terminal dimeric interfaces supported 2-fold greater KS-CP interactions, it did not increase the partial product occupancy of the homodimeric protein. Our findings cast light on molecular details of natural antibiotic biosynthesis that will aid in the design of artificial megasynth(et)ases with untold product structures and bioactivities.
Organizational Affiliation:
Department of Pharmacology and Pharmaceutical Sciences, University of Southern California, Los Angeles, CA 90089, USA.