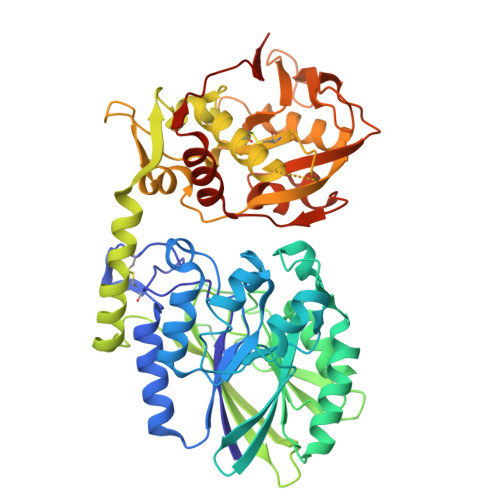
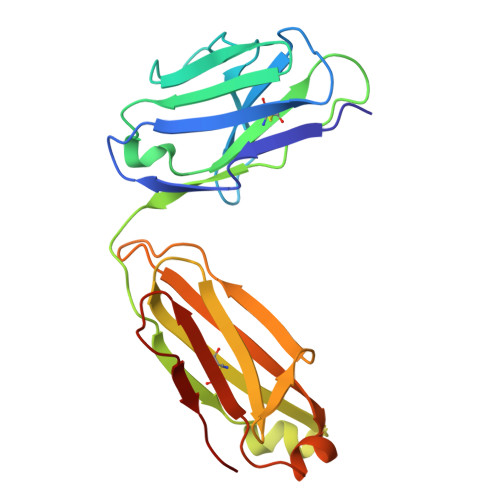
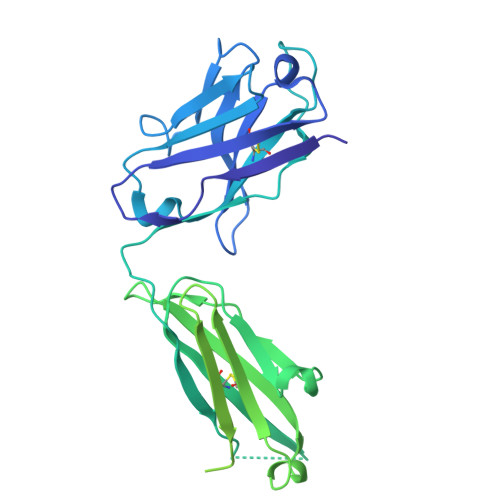
Sym024 interacts with a unique epitope on the CD73 homodimer, favoring effective bivalent binding to improve anti-PD1 therapy.
Jakobsen, J.S., Grandal, M.M., Hansen, R.W., Bansia, H., Armbruster, E., Theret, I., Skartved, N.J.O., Hald, R., Melander, M.C., Worsaae, A., Riva, M., Reckzeh, K., Vuillard, L., Lantto, J., des Georges, A., Frohlich, C.(2026) Clin Cancer Res
- PubMed: 41511395
- DOI: https://doi.org/10.1158/1078-0432.CCR-25-2406
- Primary Citation of Related Structures:
9P1M - PubMed Abstract:
Adenosine signaling may be a central immune suppressive mechanism in several cancers, and blockade of the rate-limiting CD73 AMP-to-adenosine enzyme has been demonstrated to improve clinical efficacy of PD(L)-1 immune therapy. However, deep inhibition of CD73 activity could prove difficult in tumor environments with a constant AMP supply and high CD73 levels. Here, we sought to identify, characterize, and benchmark a novel antagonistic anti-CD73 antibody, Sym024 (S95024), and to structurally decode its mode of action. Sym024, selected via functional antibody repertoire screening, was tested against benchmark anti-CD73 antibodies in primary cell, cell line in vitro binding, CD73 enzymatic activity, and T cell activation assays. Its in vivo tumor growth inhibition was examined in transplanted human or mouse tumors in immunocompetent or immunodeficient mice, and intra-tumoral enzymatic inhibition and immune cell recruitment were assessed. We investigated Sym024-CD73 interaction using surface plasmon resonance, cryo-electron microscopy, site-directed mutagenesis, and population level complex formation through size-exclusion-chromatography with light scatter mass detection. Preclinical safety and pharmacokinetics were assessed in monkeys. Sym024 effectively blocked CD73 across a large range of enzyme expression levels, comparing favorably to benchmark anti-CD73 antibodies; it improved the efficacy of PD-1 blockade in vitro as well as in vivo. Our structural data indicate that a unique one-to-one Sym024-CD73 interaction engenders this comprehensive inhibition. No pre-clinical safety flags were observed, and the pharmacokinetics profile of Sym024 supported a standard clinical dosing regimen. The comprehensive CD73 inhibition exhibited by Sym024 may improve the efficacy of anti-PD(L)-1/anti-CD73 combination treatment.
- Servier Symphogen A/S Ballerup Denmark.
Organizational Affiliation: