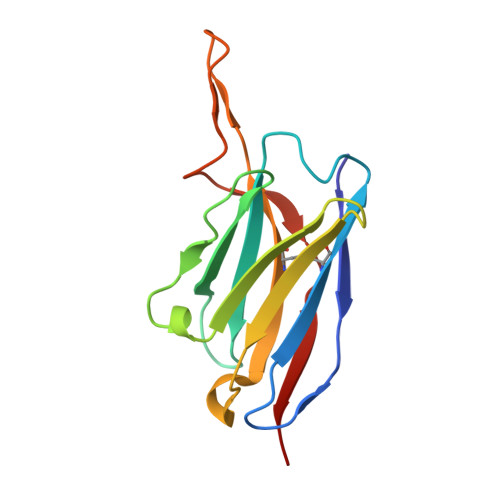
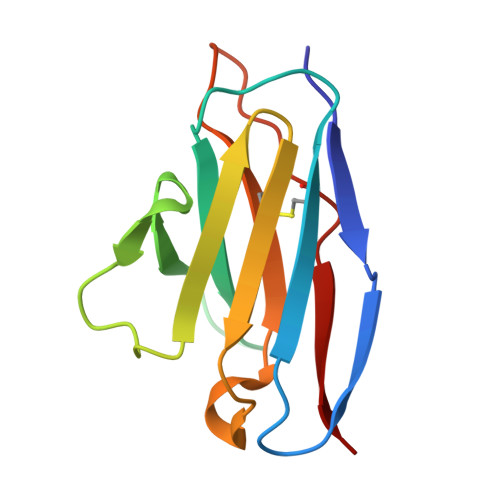
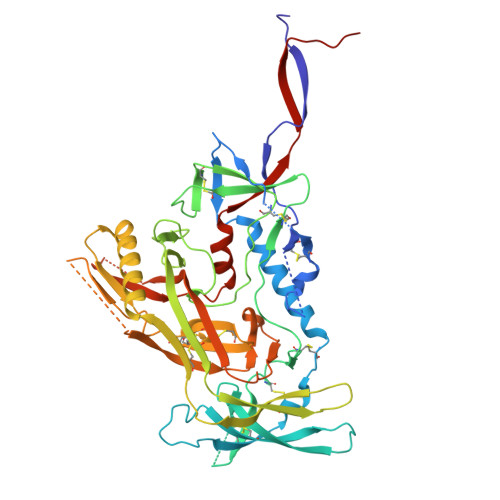
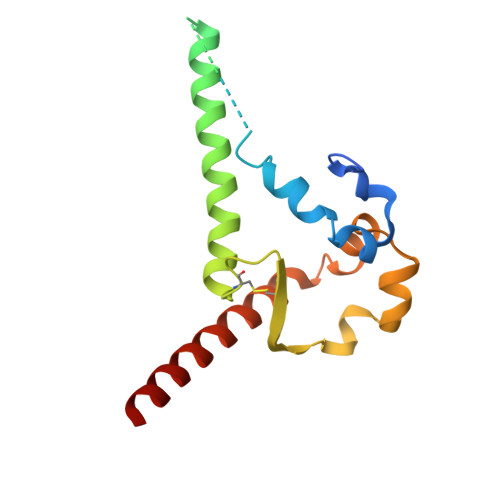
Rapid elicitation of neutralizing Asn332-glycan-independent antibodies to the V3-glycan epitope of HIV-1 Env in nonhuman primates.
Relano-Rodriguez, I., Du, J., Lin, Z.J., Kerwin, M., Tarquis-Medina, M., Urbano, E., Cui, J., Watkins, M., Lavine, C.L., Zhao, P., Habib, R., Agostino, C., Ghosh, S., Park, J., Boroughs, C., Walsh, A.A., Melo, M.B., Shukla, N., Shaw, G.M., Hahn, B.H., Irvine, D.J., Wells, L., Weiner, D.B., Seaman, M.S., Kulp, D.W., Veazey, R.S., Pallesen, J., Escolano, A.(2026) Nat Immunol
- PubMed: 41634485
- DOI: https://doi.org/10.1038/s41590-025-02408-z
- Primary Citation of Related Structures:
9O8M, 9OED - PubMed Abstract:
Sequential immunization is a promising approach to elicit broadly neutralizing antibodies (bNAbs) against the HIV-1 Envelope (Env). However, available protocols are inefficient and involve multiple immunizations over long periods of time. Here, we present WIN332, a new engineered Env immunogen that induces a new class of Asn332-glycan-independent antibodies to the conserved V3-glycan epitope of Env with low inhibitory activity indicative of a neutralization activity after a single bolus immunization in nonhuman primates. WIN332 binds to precursors of canonical human Asn332-glycan-dependent (type-I) V3-glycan bNAbs but also of a first-of-its-class Asn332-glycan-independent (type-II) V3-glycan bNAb. A single immunization elicits low inhibitory serum and monoclonal antibodies that are boosted and affinity matured with a heterologous immunogen. Electron microscopy polyclonal epitope mapping analysis of serum antibodies, antibody cloning and cryogenic electron microscopy analysis reveals that WIN332 elicits Asn332-glycan-independent antibodies with striking sequence and binding similarities with the most potent human type-I and type-II V3-glycan bNAbs. Thus, WIN332 is a promising vaccine candidate to streamline V3-glycan bNAb elicitation.
- Vaccine & Immunotherapy Center, The Wistar Institute, Philadelphia, PA, USA.
Organizational Affiliation: