Unspecific peroxygenase from Psathyrella aberdarensis
Fernandez-Garcia, A., Sanz-Aparicio, J.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Heme-thiolate peroxidase | 335 | Candolleomyces aberdarensis | Mutation(s): 0 Gene Names: EST38_g8238 EC: 1.11.2.1 | 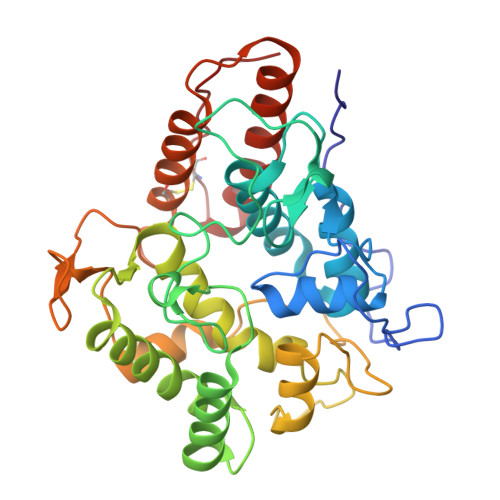 | |
UniProt | |||||
Find proteins for A0A4V1Q376 (Candolleomyces aberdarensis) Explore A0A4V1Q376 Go to UniProtKB: A0A4V1Q376 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A4V1Q376 | ||||
Glycosylation | |||||
| Glycosylation Sites: 2 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | D, F | 5 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G22768VO GlyCosmos: G22768VO GlyGen: G22768VO | |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | E | 7 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G55220VL GlyCosmos: G55220VL GlyGen: G55220VL | |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | G | 6 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G09724ZC GlyCosmos: G09724ZC GlyGen: G09724ZC | |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | H | 4 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G81315DD GlyCosmos: G81315DD GlyGen: G81315DD | |||||
| Ligands 6 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| HEM (Subject of Investigation/LOI) Query on HEM | CA [auth C], J [auth A], U [auth B] | PROTOPORPHYRIN IX CONTAINING FE C34 H32 Fe N4 O4 KABFMIBPWCXCRK-RGGAHWMASA-L |  | ||
| EPE Query on EPE | K [auth A], N [auth A], R [auth B] | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID C8 H18 N2 O4 S JKMHFZQWWAIEOD-UHFFFAOYSA-N |  | ||
| FUX (Subject of Investigation/LOI) Query on FUX | AA [auth C] BA [auth C] L [auth A] M [auth A] S [auth B] | 5-HYDROXYMETHYL-FURFURAL C6 H6 O3 NOEGNKMFWQHSLB-UHFFFAOYSA-N |  | ||
| SO4 Query on SO4 | EA [auth C], FA [auth C], Q [auth A], W [auth B], X [auth B] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| GOL Query on GOL | GA [auth C], HA [auth C], O [auth A], Y [auth B], Z [auth B] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| MG Query on MG | DA [auth C], P [auth A], V [auth B] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 272.213 | α = 90 |
| b = 75.112 | β = 111.68 |
| c = 105.385 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| XDS | data reduction |
| XDS | data scaling |
| REFMAC | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Spanish Ministry of Science, Innovation, and Universities | Spain | Synergy CAM Project Y2018/BIO-4738-EVOQUIMERA-CM |