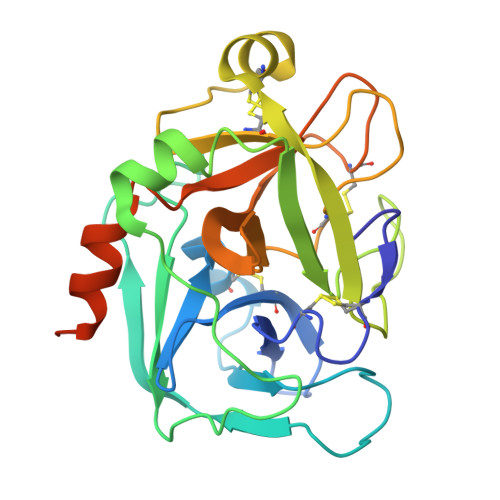
A 3.3- angstrom cryo-EM structure of an engineered high-affinity human prothrombinase complex.
Ustok, F.I., Faille, A., Huntington, J.A.(2025) Blood
- PubMed: 41144779
- DOI: https://doi.org/10.1182/blood.2025031527
- Primary Citation of Related Structures:
9I24, 9I2H - PubMed Abstract:
Thrombin is generated from prothrombin through cleavage at two sites by the enzyme prothrombinase, composed of factor (f) Xa and fVa. The affinity of fXa for fVa is low, with assembly and function dependent on phospholipid (PL) membranes. Some snakes have evolved venom versions of fXa that bind to fVa with high affinity and efficiently activate prothrombin in the absence of PL. We created a similar high-affinity, PL-independent human prothrombinase with 17 mutations to human fXa (M17). The increase in affinity enabled cryogenic electron microscopy (cryo-EM) structure determination of M17-prothrombinase to a resolution of 3.3 Å. All protein domains were well resolved in the map, except for the Gla domain of fXa. The main contacts involve the serine protease and EGF2 domains of fXa and the A2 and A3 domains of fVa, resulting in the burying of a total surface area of 4,900 Å2. The map is of sufficient quality to resolve side chain interactions, including several key M17 mutations. To aid in the placement of the loop C-terminal to the A2 domain (a2-loop), we solved a high-resolution crystal structure of fXa in complex with a synthetic a2-peptide. The acidic a2-loop interacts with the basic heparin binding site of fXa, involving a conserved antiparallel b-strand interaction. The M17-prothrombinase structure is compatible with data from biochemical and mutagenesis research, and provides important new insights into the assembly and function of the prothrombinase complex.
- University of Cambridge, Cambridge, United Kingdom.
Organizational Affiliation: