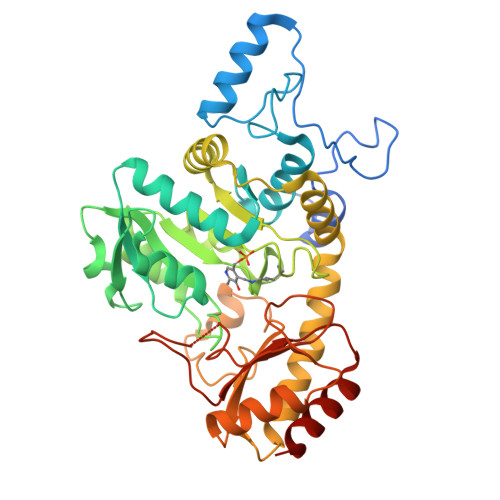
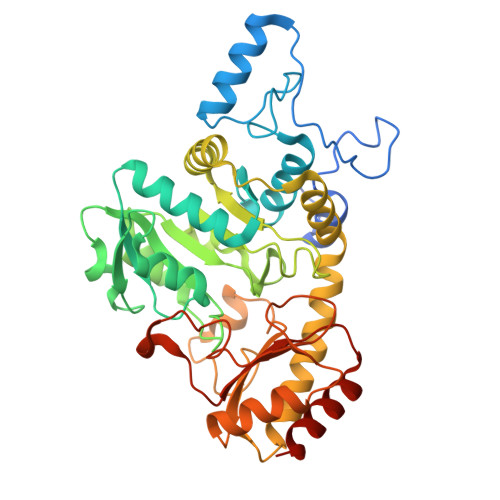
Functional and structural characterization of methionine gamma-lyases from Brevibacterium bacteria: insights into cofactor retention
Ferchaud, N., Boyer, A., Odegard, B., Hentati, S., Simonson, T., Machover, D., Kopecny, D., Briozzo, P.To be published.