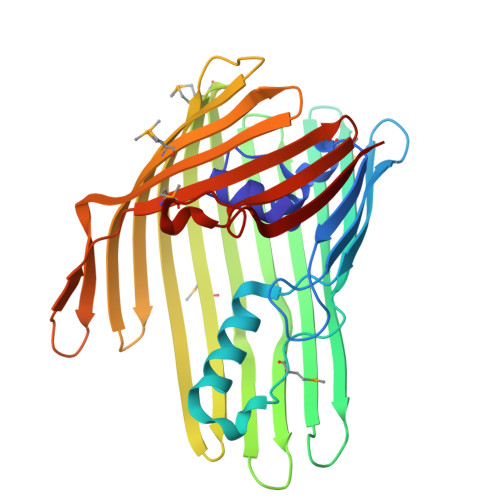
A porin-like protein used by bacterial predators defines a wider lipid-trapping superfamily.
Parr, R.J., Santin, Y.G., Ratkeviciute, G., Caulton, S.G., Radford, P., Gurvic, D., Jenkins, M., Doyle, M.T., Mead, L., Silale, A., van den Berg, B., Knowles, T.J., Sockett, R.E., Stansfeld, P.J., Laloux, G., Lovering, A.L.(2025) Nat Commun 16: 6213-6213
- PubMed: 40617869
- DOI: https://doi.org/10.1038/s41467-025-61633-0
- Primary Citation of Related Structures:
9GA1, 9GF0 - PubMed Abstract:
Outer membrane proteins (OMPs) define the surface biology of Gram-negative bacteria, with roles in adhesion, transport, catalysis and signalling. Specifically, porin beta-barrels are common diffusion channels, predominantly monomeric/trimeric in nature. Here we show that the major OMP of the bacterial predator Bdellovibrio bacteriovorus, PopA, differs from this architecture, forming a pentameric porin-like superstructure. Our X-ray and cryo-EM structures reveal a bowl-shape composite outer β-wall, which houses a central chamber that encloses a section of the lipid bilayer. We demonstrate that PopA, reported to insert into prey inner membrane, causes defects when directed into Escherichia coli membranes. We discover widespread PopA homologues, including likely tetramers and hexamers, that retain the lipid chamber; a similar chamber is formed by an unrelated smaller closed-barrel family, implicating this as a general feature. Our work thus defines oligomeric OMP superfamilies, whose deviation from prior structures requires us to revisit existing membrane-interaction motifs and folding models.
- School of Biosciences, University of Birmingham, Birmingham, UK.
Organizational Affiliation: