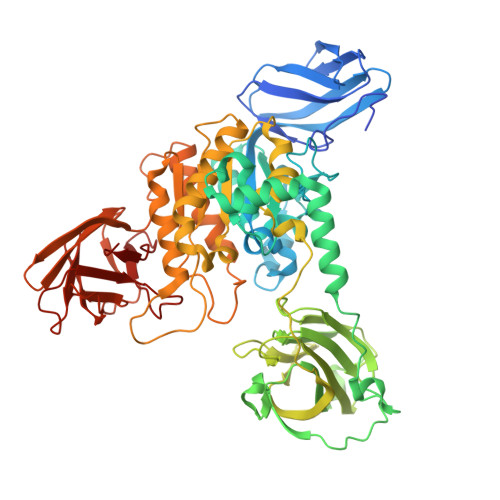
Insights into a water-mediated catalytic triad architecture in CE20 carbohydrate esterases.
Teune, M., Vieira, P.S., Dohler, T., Palm, G.J., Dutschei, T., Bartosik, D., Berndt, L., Persinoti, G.F., Maass, S., Becher, D., Schweder, T., Murakami, M.T., Lammers, M., Bornscheuer, U.T.(2025) Nat Commun 16: 7034-7034
- PubMed: 40745183
- DOI: https://doi.org/10.1038/s41467-025-62387-5
- Primary Citation of Related Structures:
9EGA, 9H4U - PubMed Abstract:
Carbohydrate esterases modify polysaccharides by removing different ester moieties thereby affecting their physicochemical properties and their accessibility by glycoside hydrolases. We determined the full-length structures of two members (Fl8CE20_II and PpCE20_II) from the carbohydrate esterase family 20 (CE20) by X-ray crystallography that feature an ancillary domain, inserted into the catalytic SGNH-hydrolase domain. Detailed structural analysis identifies a so far undescribed catalytic triad architecture which lacks the typical aspartate for polarization of the histidine but instead reveals a precisely coordinated water molecule mediating contact between the His and Asp. This coordinated water in the Ser-His-(H 2 O-Asp/Asn) motif, as further confirmed by mutational studies and by determination of kinetic constants, is crucial for catalytic activity. We therefore term this active site architecture a water-mediated catalytic triad.
- Department of Biotechnology & Enzyme Catalysis, Institute of Biochemistry, University Greifswald, Greifswald, Germany.
Organizational Affiliation: