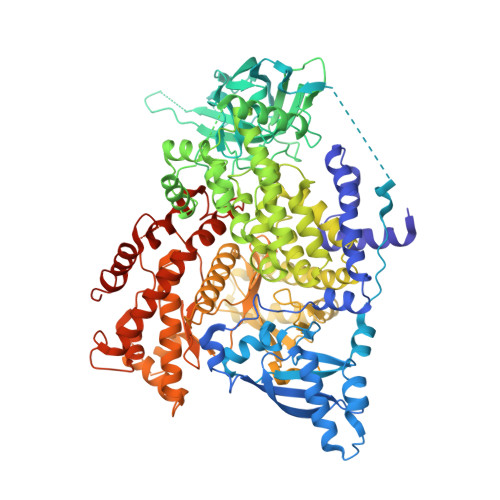
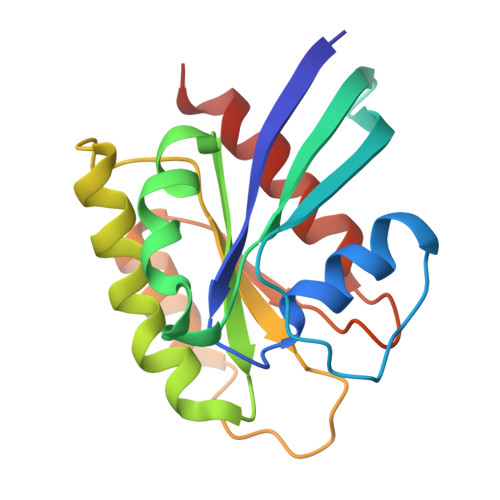
Molecular glues that facilitate RAS binding to PI3K alpha promote glucose uptake without insulin.
Terayama, K., Furuzono, S., Fer, N., Yan, W., Young, L.C., Czyzyk, D.J., Goldstein de Salazar, R., Sasaki, M., Uozumi, A., Konishi, M., Kanda, S., Sogawa, Y., Yamaguchi, M., Tsuji, T., Kuroyanagi, J., Hayashi, M., Ogura, Y., Simanshu, D.K., Kubota, K., Tanaka, J., McCormick, F.(2025) Science 389: 402-408
- PubMed: 40705882
- DOI: https://doi.org/10.1126/science.adr9097
- Primary Citation of Related Structures:
9CMK, 9CML, 9CMV - PubMed Abstract:
While exploring strategies to control blood glucose concentrations in diabetes, we identified so-called molecular glues D223 and D927 that promote glucose uptake in the absence of insulin. They act by increasing the binding affinity of phosphoinositide 3-kinase α (PI3Kα) catalytic subunit p110α to canonical small guanosine triphosphatase RAS proteins and to RRAS, RRAS2, and MRAS by three orders of magnitude. The compounds bind to the RAS-binding domain of p110α, stabilizing the secondary structures of the PI3Kα in a RAS-binding conformation and forming direct interactions with RAS residues tyrosine-40 and arginine-41. In vivo, D927 mimicked the effects of insulin: It rapidly lowered blood glucose concentrations, enhanced glucose metabolism in normal and Zucker fatty rats, and improved hyperglycemia in models of type 1 and type 2 diabetes, even in insulin-deficient diabetic animals.
- Cardiovascular Metabolic Research Laboratories, Daiichi Sankyo Co., Ltd., Tokyo, Japan.
Organizational Affiliation: