c-KIT kinase domain in complex with crenolanib
Teuber, A., Niggenaber, J., Kleinboelting, S.B., Landel, I., Warmuth, J., Busick, D., Mueller, M.P., Bauer, S., Rauh, D.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Mast/stem cell growth factor receptor Kit | 327 | Homo sapiens | Mutation(s): 14 Gene Names: KIT, SCFR EC: 2.7.10.1 | 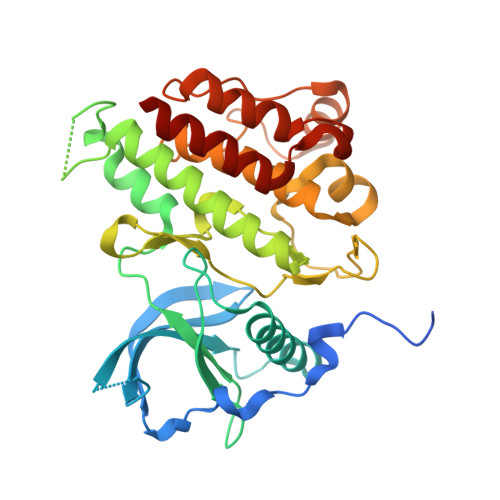 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P10721 (Homo sapiens) Explore P10721 Go to UniProtKB: P10721 | |||||
PHAROS: P10721 GTEx: ENSG00000157404 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P10721 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| 6T2 (Subject of Investigation/LOI) Query on 6T2 | C [auth A], D [auth B] | 1-(2-{5-[(3-Methyloxetan-3-yl)methoxy]-1H-benzimidazol-1-yl}quinolin-8-yl)piperidin-4-amine C26 H29 N5 O2 DYNHJHQFHQTFTP-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 59.54 | α = 90 |
| b = 59.53 | β = 90 |
| c = 191.41 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PDB_EXTRACT | data extraction |
| PHENIX | refinement |
| XDS | data reduction |
| XSCALE | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| German Research Foundation (DFG) | Germany | RA 1055/3-2 |
| European Regional Development Fund | European Union | EFRE-800400 |
| German Federal Ministry for Education and Research | Germany | 01ZX2201B |
| Other government | NW21-062C | |
| Other private | Ex-2021-0033 |