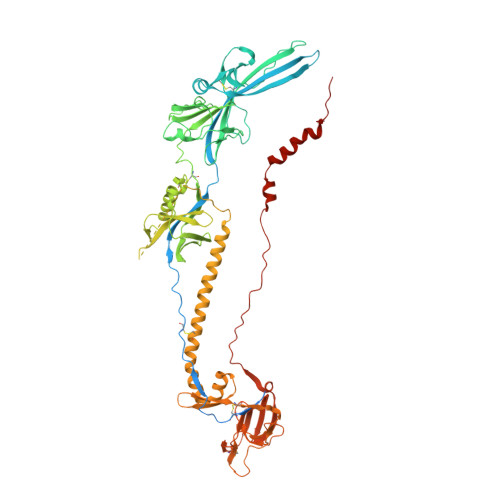
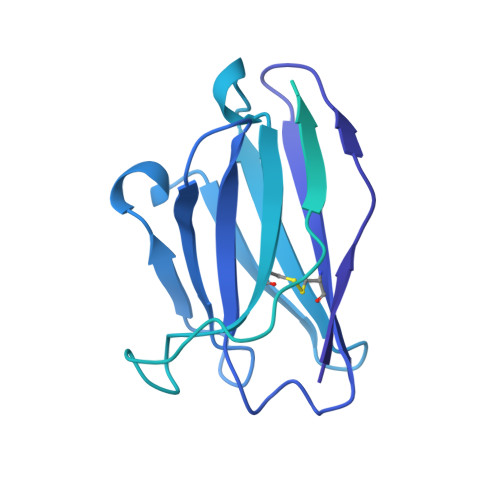
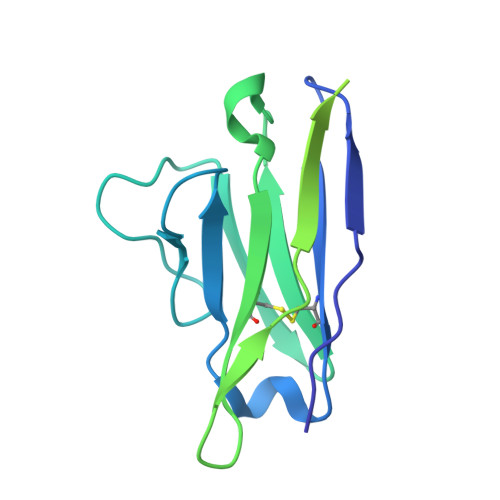
Development of a highly effective combination monoclonal antibody therapy against Herpes simplex virus.
Seyfizadeh, N., Kalbermatter, D., Imhof, T., Ries, M., Muller, C., Jenner, L., Blumenschein, E., Yendrzheyevskiy, A., Grun, F., Moog, K., Eckert, D., Engel, R., Diebolder, P., Chami, M., Krauss, J., Schaller, T., Arndt, M.(2024) J Biomed Sci 31: 56-56
- PubMed: 38807208
- DOI: https://doi.org/10.1186/s12929-024-01045-2
- Primary Citation of Related Structures:
8RGZ, 8RH0, 8RH1, 8RH2 - PubMed Abstract:
Infections with Herpes simplex virus (HSV)-1 or -2 usually present as mild chronic recurrent disease, however in rare cases can result in life-threatening conditions with a large spectrum of pathology. Monoclonal antibody therapy has great potential especially to treat infections with virus resistant to standard therapies. HDIT101, a humanized IgG targeting HSV-1/2 gB was previously investigated in phase 2 clinical trials. The aim of this study was to develop a next-generation therapy by combining different antiviral monoclonal antibodies. A lymph-node derived phage display library (LYNDAL) was screened against recombinant gB from Herpes simplex virus (HSV) -1 and HDIT102 scFv was selected for its binding characteristics using bio-layer interferometry. HDIT102 was further developed as fully human IgG and tested alone or in combination with HDIT101, a clinically tested humanized anti-HSV IgG, in vitro and in vivo. T-cell stimulating activities by antigen-presenting cells treated with IgG-HSV immune complexes were analyzed using primary human cells. To determine the epitopes, the cryo-EM structures of HDIT101 or HDIT102 Fab bound to HSV-1F as well as HSV-2G gB protein were solved at resolutions < 3.5 Å. HDIT102 Fab showed strong binding to HSV-1F gB with Kd of 8.95 × 10 -11 M and to HSV-2G gB with Kd of 3.29 × 10 -11 M. Neutralization of cell-free virus and inhibition of cell-to-cell spread were comparable between HDIT101 and HDIT102. Both antibodies induced internalization of gB from the cell surface into acidic endosomes by binding distinct epitopes in domain I of gB and compete for binding. CryoEM analyses revealed the ability to form heterogenic immune complexes consisting of two HDIT102 and one HDIT101 Fab bound to one gB trimeric molecule. Both antibodies mediated antibody-dependent phagocytosis by antigen presenting cells which stimulated autologous T-cell activation. In vivo, the combination of HDIT101 and HDIT102 demonstrated synergistic effects on survival and clinical outcome in immunocompetent BALB/cOlaHsd mice. This biochemical and immunological study showcases the potential of an effective combination therapy with two monoclonal anti-gB IgGs for the treatment of HSV-1/2 induced disease conditions.
Organizational Affiliation:
Heidelberg ImmunoTherapeutics GmbH, Max-Jarecki Str. 21, Heidelberg, 69115, Germany.