RAMD identification of substrate binding pathways to the active site of dUTPase
Toth, J., Vertessy, B.G., Leveles, I., Bendes, A., Lopata, A.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Deoxyuridine 5'-triphosphate nucleotidohydrolase | 174 | Mycobacterium tuberculosis | Mutation(s): 1 Gene Names: dut, Rv2697c, MT2771, MTCY05A6.18c EC: 3.6.1.23 | 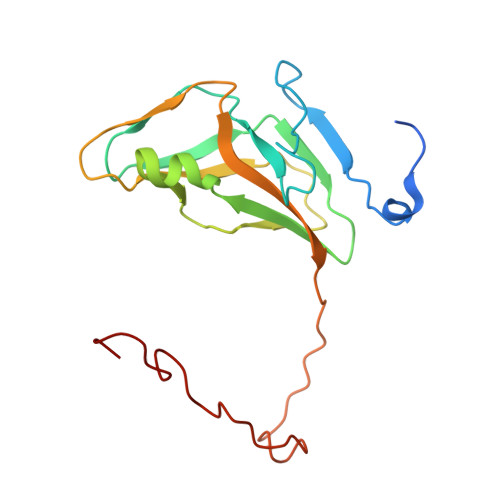 | |
UniProt | |||||
Find proteins for P9WNS5 (Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv)) Explore P9WNS5 Go to UniProtKB: P9WNS5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P9WNS5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| DUP Query on DUP | B [auth A] | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE C9 H16 N3 O13 P3 XZLLMTSKYYYJLH-SHYZEUOFSA-N |  | ||
| TRS Query on TRS | C [auth A] | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL C4 H12 N O3 LENZDBCJOHFCAS-UHFFFAOYSA-O |  | ||
| GOL Query on GOL | E [auth A] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| MG Query on MG | D [auth A] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 54.94 | α = 90 |
| b = 54.94 | β = 90 |
| c = 83.86 | γ = 120 |
| Software Name | Purpose |
|---|---|
| CrystalClear | data collection |
| MOLREP | phasing |
| REFMAC | refinement |
| MOSFLM | data reduction |
| SCALA | data scaling |