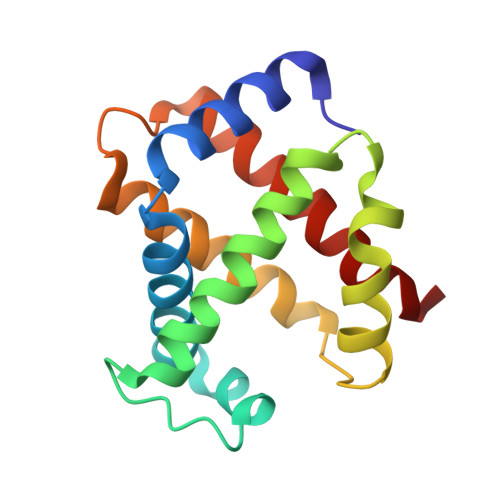
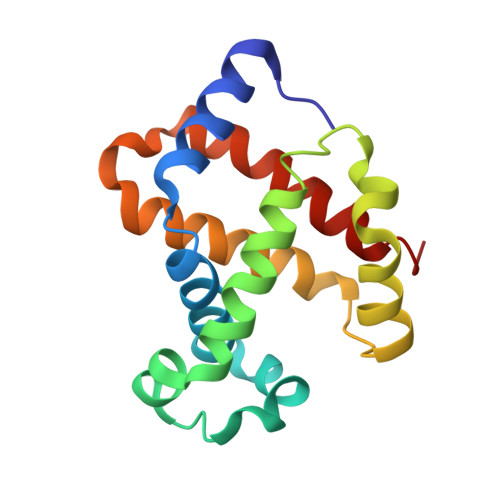
Structure of relaxed-state human hemoglobin: insight into ligand uptake, transport and release.
Jenkins, J.D., Musayev, F.N., Danso-Danquah, R., Abraham, D.J., Safo, M.K.(2009) Acta Crystallogr D Biol Crystallogr 65: 41-48
- PubMed: 19153465
- DOI: https://doi.org/10.1107/S0907444908037256
- Primary Citation of Related Structures:
3D17 - PubMed Abstract:
Hemoglobin was one of the first protein structures to be determined by X-ray crystallography and served as a basis for the two-state MWC model for the mechanism of allosteric proteins. Since then, there has been an ongoing debate about whether Hb allostery involves the unliganded tense T state and the liganded relaxed R state or whether it involves the T state and an ensemble of liganded relaxed states. In fact, the former model is inconsistent with many functional observations, as well as the recent discoveries of several relaxed-state Hb structures such as RR2, R3 and R2. One school of thought has suggested the R2 state to be the physiologically relevant relaxed end state, with the R state mediating the T-->R2 transition. X-ray studies have been performed on human carbonmonoxy Hb at a resolution of 2.8 A. The ensuing liganded quaternary structure is different from previously reported liganded Hb structures. The distal beta-heme pocket is the largest when compared with other liganded Hb structures, partly owing to rotation of betaHis63(E7) out of the distal pocket, creating a ligand channel to the solvent. The structure also shows unusually smaller alpha- and beta-clefts. Results from this study taken in conjunction with previous findings suggest that multiple liganded Hb states with different quaternary structures may be involved in ligand uptake, stabilization, transport and release.
- Department of Medicinal Chemistry, School of Pharmacy and Institute for Structural Biology and Drug Discovery, Virginia Commonwealth University, Richmond, VA 23219, USA.
Organizational Affiliation: