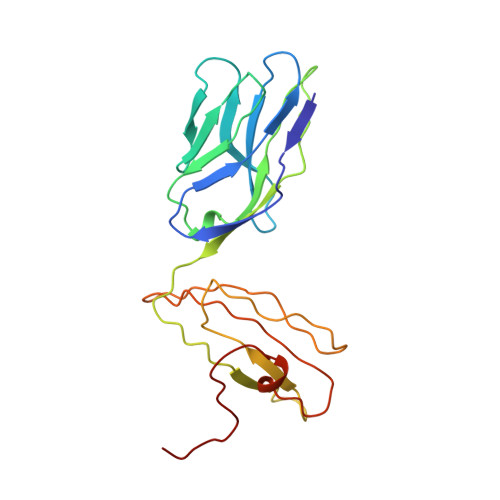
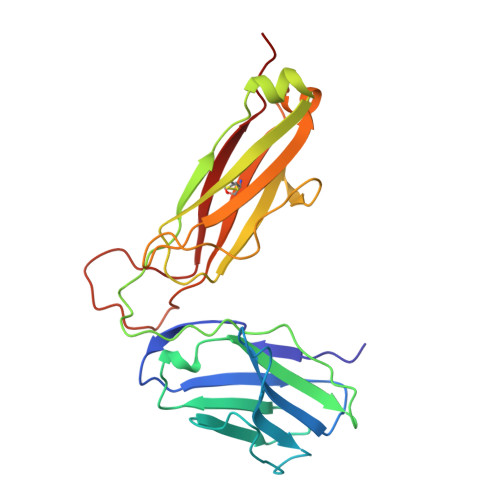
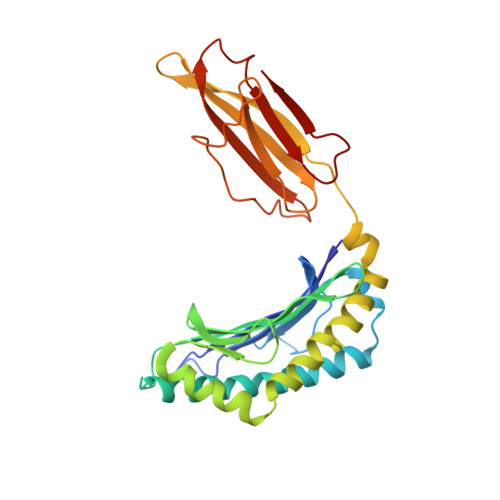
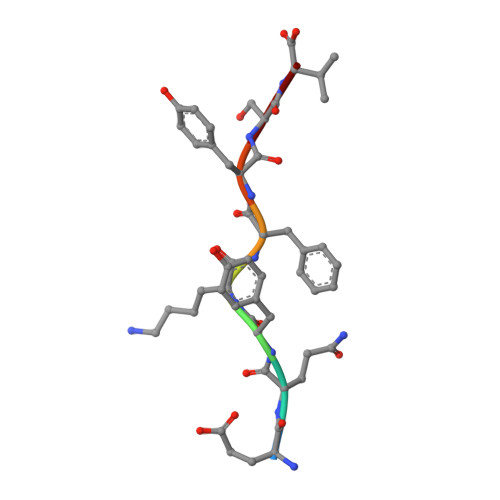
Structural basis of plasticity in T cell receptor recognition of a self peptide-MHC antigen.
Garcia, K.C., Degano, M., Pease, L.R., Huang, M., Peterson, P.A., Teyton, L., Wilson, I.A.(1998) Science 279: 1166-1172
- PubMed: 9469799
- DOI: https://doi.org/10.1126/science.279.5354.1166
- Primary Citation of Related Structures:
2CKB - PubMed Abstract:
The T cell receptor (TCR) inherently has dual specificity. T cells must recognize self-antigens in the thymus during maturation and then discriminate between foreign pathogens in the periphery. A molecular basis for this cross-reactivity is elucidated by the crystal structure of the alloreactive 2C TCR bound to self peptide-major histocompatibility complex (pMHC) antigen H-2Kb-dEV8 refined against anisotropic 3.0 angstrom resolution x-ray data. The interface between peptide and TCR exhibits extremely poor shape complementarity, and the TCR beta chain complementarity-determining region 3 (CDR3) has minimal interaction with the dEV8 peptide. Large conformational changes in three of the TCR CDR loops are induced upon binding, providing a mechanism of structural plasticity to accommodate a variety of different peptide antigens. Extensive TCR interaction with the pMHC alpha helices suggests a generalized orientation that is mediated by the Valpha domain of the TCR and rationalizes how TCRs can effectively "scan" different peptides bound within a large, low-affinity MHC structural framework for those that provide the slight additional kinetic stabilization required for signaling.
- Department of Molecular Biology and the Skaggs Institute of Chemical Biology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037, USA.
Organizational Affiliation: