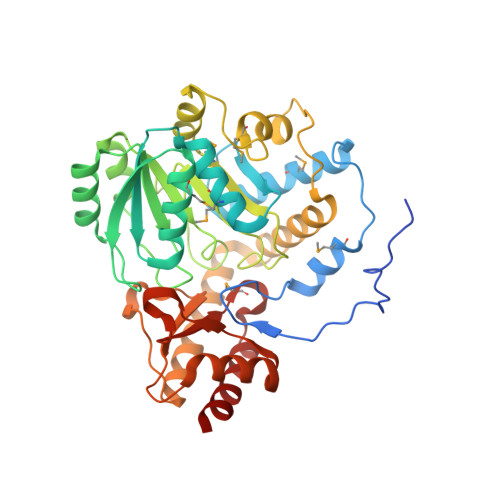
Crystal structure of an alanine-glyoxylate aminotransferase from Anabaena sp. at 1.70 A resolution reveals a noncovalently linked PLP cofactor
Han, G.W., Schwarzenbacher, R., Page, R., Jaroszewski, L., Abdubek, P., Ambing, E., Biorac, T., Canaves, J.M., Chiu, H.J., Dai, X., Deacon, A.M., DiDonato, M., Elsliger, M.A., Godzik, A., Grittini, C., Grzechnik, S.K., Hale, J., Hampton, E., Haugen, J., Hornsby, M., Klock, H.E., Koesema, E., Kreusch, A., Kuhn, P., Lesley, S.A., Levin, I., McMullan, D., McPhillips, T.M., Miller, M.D., Morse, A., Moy, K., Nigoghossian, E., Ouyang, J., Paulsen, J., Quijano, K., Reyes, R., Sims, E., Spraggon, G., Stevens, R.C., van den Bedem, H., Velasquez, J., Vincent, J., von Delft, F., Wang, X., West, B., White, A., Wolf, G., Xu, Q., Zagnitko, O., Hodgson, K.O., Wooley, J., Wilson, I.A.(2005) Proteins 58: 971-975
- PubMed: 15657930
- DOI: https://doi.org/10.1002/prot.20360
- Primary Citation of Related Structures:
1VJO - The Joint Center for Structural Genomics, The Scripps Research Institute, La Jolla, CA 92037, USA.
Organizational Affiliation: