News
Paper Published: 3DEM Structure-Map Validation Recommendations
08/18 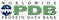
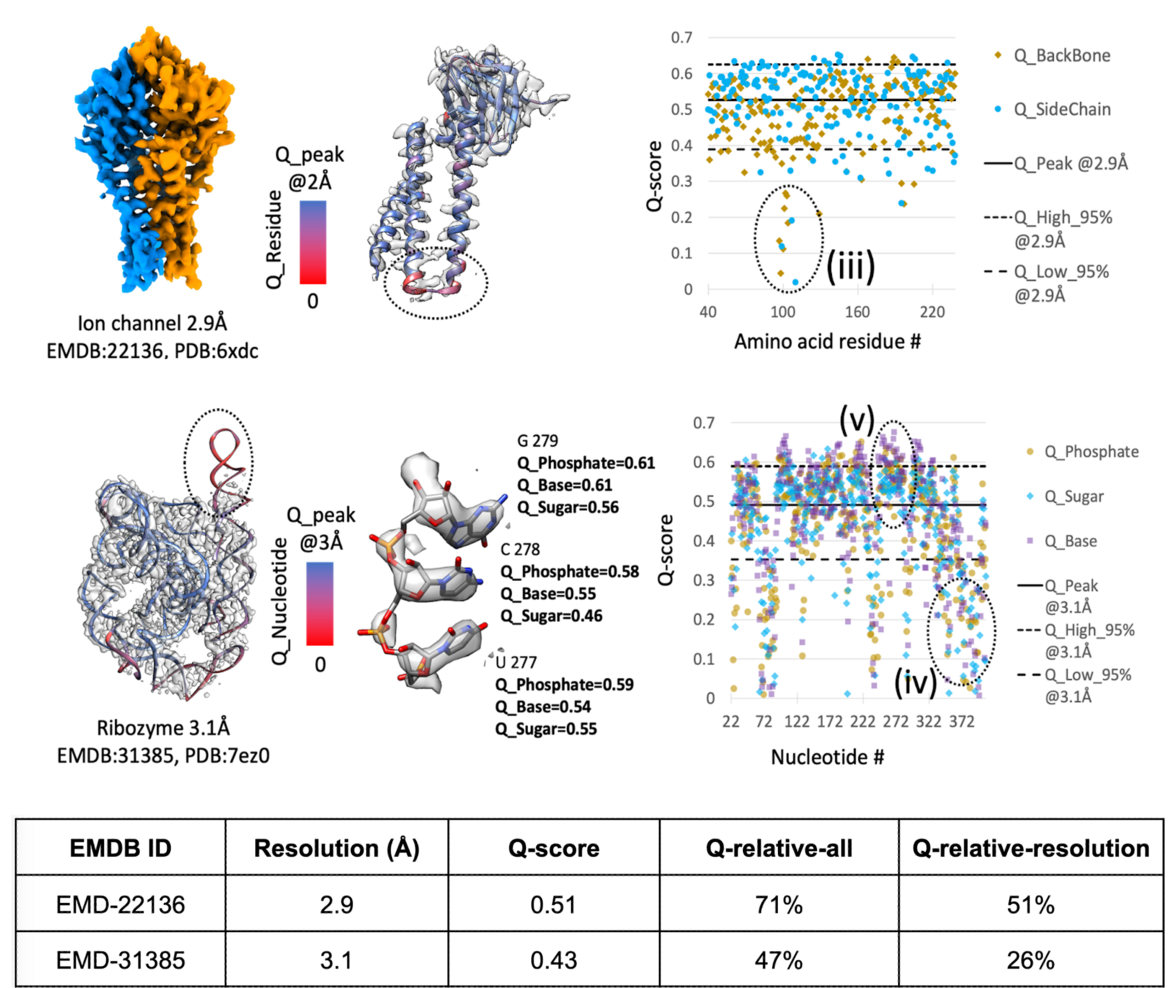
A new publication describes two percentile-based metrics: Q_relative_all and Q_relative_resolution. The publication presents a statistical analysis of Q-score and these new percentile based validation metrics.
Q_relative_all is a single value percentile, representing the overall quality of the model-map fitness. It is derived by comparing a model-map average Q-score to all model-map average Q-scores calculated for 3DEM structures in the PDB/EMDB. Higher Q_relative_all percentile values, infer higher quality in the model-map fitness.
Q_relative_resolution is a single value percentile, derived by comparing the average Q-score of a model-map with the Q-scores of other model-map entries of similar resolution. This metric was developed from the principle that average Q-score is correlated with map resolution. Q_relative_resolution percentile values close to 50% therefore represent a score that is commonly observed for entries in the PDB/EMDB of similar resolution, inferring model-map fitness is typical for an entry of a particular resolution. Notably low or high Q_relative_resolution percentiles may be helpful to guide further review of model-maps.
The EMDB has made these metrics available since Spring 2024 via emdatabank.org and since the publication of the paper, the wwPDB is in the process of incorporating these metrics into wwPDB validation reports by providing the Q-score percentile slider for the assessment of model-map fitness of 3DEM entries. This validation enhancement will help depositors, reviewers and the community to improve the quality of data submissions and assist users to assess 3DEM data quality in the PDB and EMDB, wwPDB core archives.
Q-score as a reliability measure for protein, nucleic acid and small-molecule atomic coordinate models derived from 3DEM maps
Grigore Pintilie, Chenghua Shao, Zhe Wang, Brian P Hudson, Justin W Flatt, Michael F Schmid, Kyle L Morris, Stephen K Burley, Wah Chiu
(2025) Acta Cryst D81: 410-422 doi: 10.1107/S2059798325005923