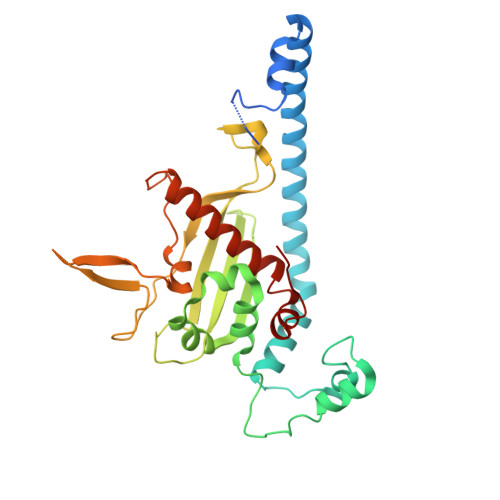
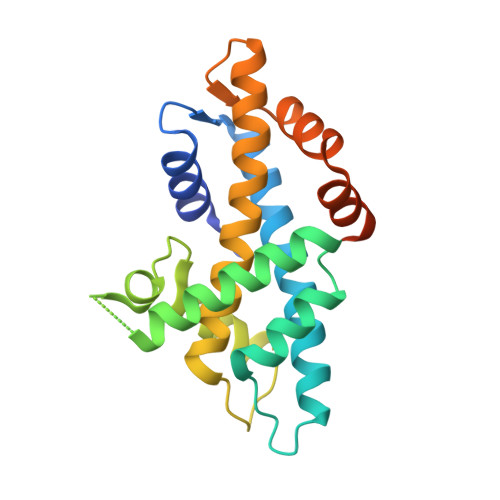
Structural insights into H2A-H2B and H2A.Z-H2B sliding on histone chaperone NAP1.
Xu, L., Zhang, J., Wang, Y., Liu, D., Zeng, C., Chen, J., Pan, X.(2025) Acta Biochim Biophys Sin (Shanghai)
- PubMed: 41403246
- DOI: https://doi.org/10.3724/abbs.2025241
- Primary Citation of Related Structures:
9LN8 - PubMed Abstract:
The evolutionarily conserved nucleosome assembly protein 1 (NAP1) functions as a histone chaperone for H2A-H2B, regulating nucleosome assembly and maintaining chromatin integrity. However, the dynamic and variable nature of the interactions between acidic NAP1 and basic H2A-H2B has obscured the molecular basis of its chaperoning activity. Here, we report the crystal structures of Caenorhabditis elegans NAP1 (CeNAP1) in complex with Xenopus laevis H2A-H2B (XlH2A-H2B) and with C . elegans H2A.Z-H2B (CeH2A.Z-H2B) at 3.35 Å and 2.8 Å, respectively. In our structures, H2A/H2A.Z-H2B binds to the acidic concave surface of CeNAP1 in three distinct poses, with two in the CeNAP1-XlH2A-H2B complex and one in the CeNAP1-CeH2A.Z-H2B complex. These poses are different from the two poses observed in the previously reported CeNAP1-CeH2A/H2A.Z-H2B structures. The predominant interaction involves engagement of the acidic CeNAP1 α6-carboxy-terminal (C-terminal) tail by the basic H2A/H2A.Z αN-α1 region, stabilized by salt bridges and electrostatic interactions. A comparative analysis of all five known poses reveals that H2A/H2A.Z-H2B can shift approximately 20.7 Å along the α6-C-terminal tail-C'-terminal tail-α6' axis. These findings demonstrate a sliding binding mode of H2A/H2A.Z-H2B on NAP1, providing new mechanistic insights into nucleosome assembly activity of histone chaperones.