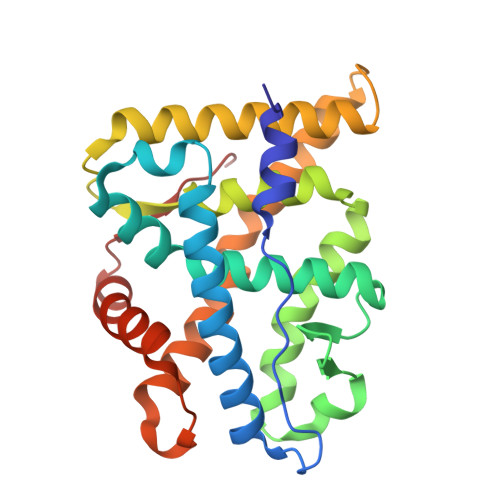
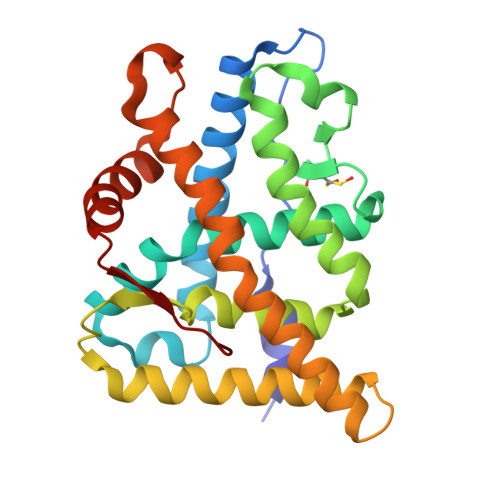
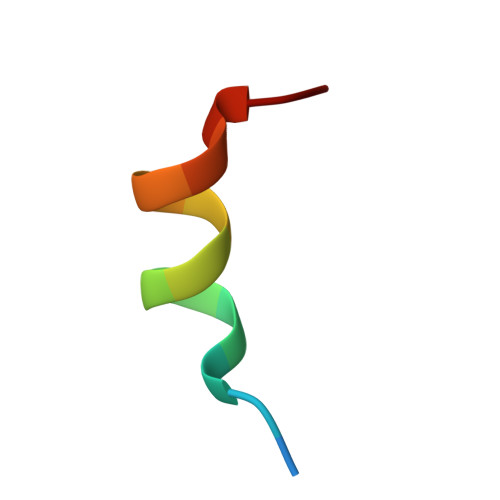The multimerization pathway of the glucocorticoid receptor.
Alegre-Marti, A., Jimenez-Panizo, A., Lafuente, A.L., Johnson, T.A., Montoya-Novoa, I., Peralta-Moreno, M.N., Montanya-Valluguera, P., Ponseti-Pons, J., Abella, M., Kim, S., Diaz, M., Vilaseca, M., Perez, P., Fernandez-Recio, J., Rubio-Martinez, J., Presman, D.M., Hager, G.L., Fuentes-Prior, P., Estebanez-Perpina, E.(2025) Nucleic Acids Res 53
- PubMed: 41118578
- DOI: https://doi.org/10.1093/nar/gkaf1003
- Primary Citation of Related Structures:
9HDF - PubMed Abstract:
The glucocorticoid receptor (GR) is a leading drug target due to its antiinflammatory and immunosuppressive roles. The functional oligomeric conformation of full-length GR (FL-GR), which is key for its biological activity, remains disputed. Here we present a new crystal structure of agonist-bound GR ligand-binding domain (GR-LBD) comprising eight copies of a noncanonical dimer. We verified the biological relevance of this dimer for receptor multimerization in wild-type and selected FL-GR mutants using molecular dynamics and crosslinking-mass spectrometry together with fluorescence microscopy and transcriptomic analysis in living cells. Self-association of this GR-LBD basic dimer in two mutually exclusive assemblies reveals clues for FL-GR multimerization and activity in cells. We propose a model for the structure of multidomain GR based on our new data and suggest a detailed oligomerization pathway. This model reconciles all currently available structural and functional information and provides a more comprehensive understanding of the rare disorder, generalized glucocorticoid resistance.
- Department of Biochemistry and Molecular Biomedicine, Faculty of Biology, University of Barcelona (UB), Barcelona 08028, Spain.
Organizational Affiliation: