Polarity protein Par6 facilitates the processive phosphorylation of Lgl via a dynamic interaction with aPKC
Almagor, L., Weis, W.I.(2025) Commun Biol
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2025) Commun Biol
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| LLGL scribble cell polarity complex component 2 | 980 | Homo sapiens | Mutation(s): 0 Gene Names: LLGL2 | 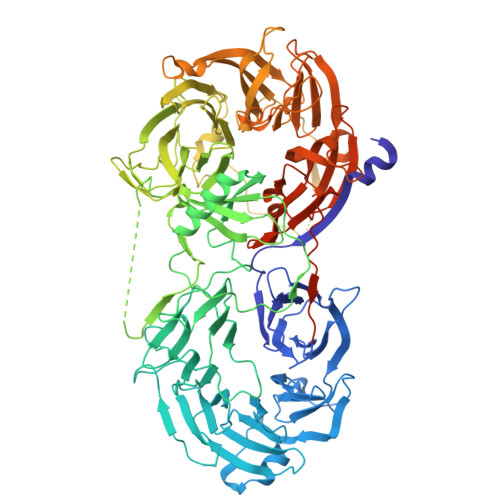 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q6P1M3 (Homo sapiens) Explore Q6P1M3 Go to UniProtKB: Q6P1M3 | |||||
PHAROS: Q6P1M3 GTEx: ENSG00000073350 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q6P1M3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Protein kinase C iota type | 596 | Homo sapiens | Mutation(s): 0 Gene Names: PRKCI, DXS1179E EC: 2.7.11.13 | 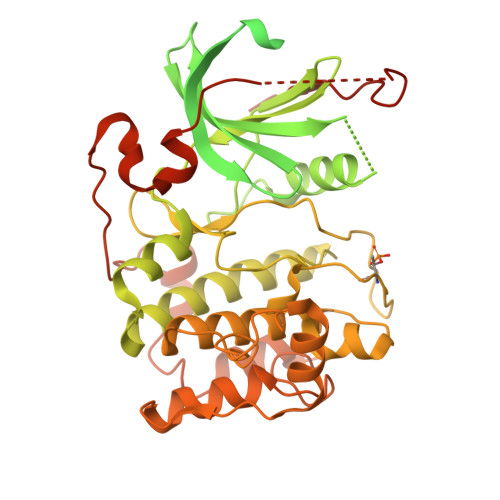 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P41743 (Homo sapiens) Explore P41743 Go to UniProtKB: P41743 | |||||
PHAROS: P41743 GTEx: ENSG00000163558 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P41743 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Partitioning defective 6 homolog beta | 383 | Mus musculus | Mutation(s): 0 Gene Names: Pard6b, Par6b | 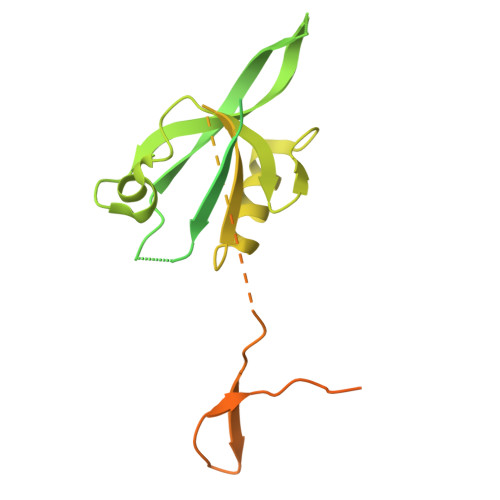 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| TPO Query on TPO | B | L-PEPTIDE LINKING | C4 H10 N O6 P |  | THR |
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | cryoSPARC |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |