Molecular mechanism of transition-state inhibitors of bacterial antibiotic efflux pumps
Boernsen, C., Mueller, R.T., Pos, K.M.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Multidrug efflux pump subunit AcrB | 1,057 | Escherichia coli K-12 | Mutation(s): 0 Gene Names: acrB, acrE, b0462, JW0451 | 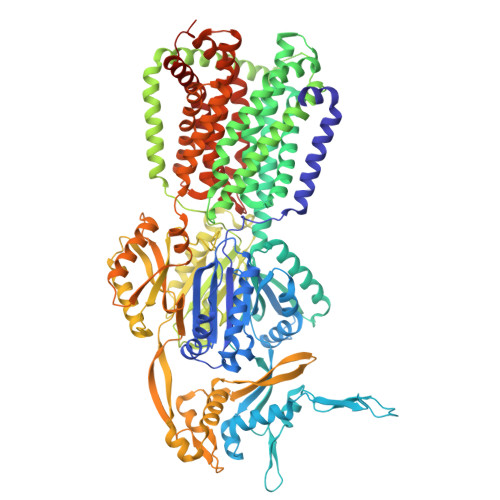 | |
UniProt | |||||
Find proteins for P31224 (Escherichia coli (strain K12)) Explore P31224 Go to UniProtKB: P31224 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P31224 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DARPIN | 169 | synthetic construct | Mutation(s): 0 | 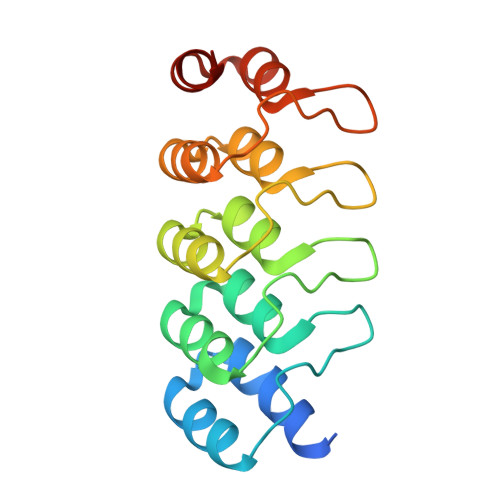 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 13 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| LMT Query on LMT | F [auth A] G [auth A] IA [auth B] J [auth A] JA [auth B] | DODECYL-BETA-D-MALTOSIDE C24 H46 O11 NLEBIOOXCVAHBD-QKMCSOCLSA-N |  | ||
| LPX Query on LPX | RB [auth C], TB [auth C] | (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate C21 H44 N O7 P YVYMBNSKXOXSKW-FQEVSTJZSA-N |  | ||
| DDR Query on DDR | NA [auth B] | (2S)-3-hydroxypropane-1,2-diyl didecanoate C23 H44 O5 GNSDEDOVXZDMKM-NRFANRHFSA-N |  | ||
| XE9 (Subject of Investigation/LOI) Query on XE9 | AA [auth A] | [3-(3-chloranyl-2-piperazin-1-yl-quinolin-6-yl)phenyl]methanamine C20 H21 Cl N4 PMBAQAZUQBDHLD-UHFFFAOYSA-N |  | ||
| DDQ Query on DDQ | OC [auth C], UA [auth B] | DECYLAMINE-N,N-DIMETHYL-N-OXIDE C12 H27 N O ZRKZFNZPJKEWPC-UHFFFAOYSA-N |  | ||
| C14 Query on C14 | I [auth A], SB [auth C] | TETRADECANE C14 H30 BGHCVCJVXZWKCC-UHFFFAOYSA-N |  | ||
| D12 Query on D12 | BB [auth B], CB [auth B], W [auth A], Y [auth A] | DODECANE C12 H26 SNRUBQQJIBEYMU-UHFFFAOYSA-N |  | ||
| D10 Query on D10 | CA [auth A], KA [auth B] | DECANE C10 H22 DIOQZVSQGTUSAI-UHFFFAOYSA-N |  | ||
| OCT Query on OCT | AC [auth C] GC [auth C] H [auth A] KB [auth B] L [auth A] | N-OCTANE C8 H18 TVMXDCGIABBOFY-UHFFFAOYSA-N |  | ||
| SO4 Query on SO4 | FC [auth C], VA [auth B], ZB [auth C] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| GOL Query on GOL | AB [auth B] BC [auth C] CC [auth C] DA [auth A] DC [auth C] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| HEX Query on HEX | BA [auth A] DB [auth B] EB [auth B] JB [auth B] MC [auth C] | HEXANE C6 H14 VLKZOEOYAKHREP-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | FB [auth B] GB [auth B] HA [auth B] HB [auth B] IB [auth B] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 146.184 | α = 90 |
| b = 161.143 | β = 90 |
| c = 244.391 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| German Federal Ministry for Education and Research | Germany | BMBF-16GW0236K |
| Agence Nationale de la Recherche (ANR) | France | -- |