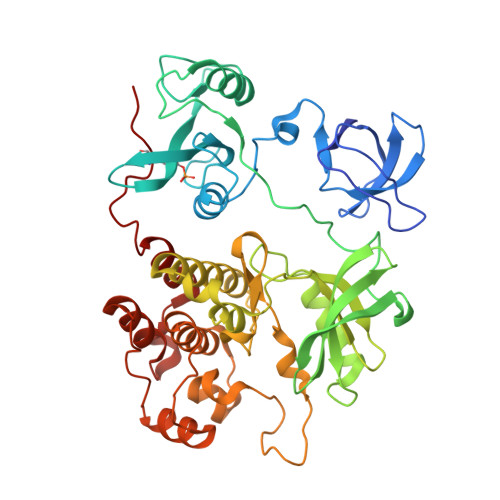
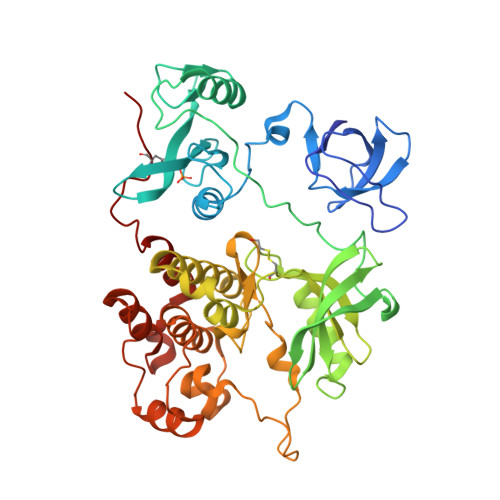
ATP-site inhibitors induce unique conformations of the acute myeloid leukemia-associated Src-family kinase, Fgr.
Du, S., Alvarado, J.J., Wales, T.E., Moroco, J.A., Engen, J.R., Smithgall, T.E.(2022) Structure 30: 1508-1517.e3
- PubMed: 36115344
- DOI: https://doi.org/10.1016/j.str.2022.08.008
- Primary Citation of Related Structures:
7UY0, 7UY3 - PubMed Abstract:
The Src-family kinase Fgr is expressed primarily in myeloid hematopoietic cells and contributes to myeloid leukemia. Here, we present X-ray crystal structures of Fgr bound to the ATP-site inhibitors A-419259 and TL02-59, which show promise as anti-leukemic agents. A-419259 induces a closed Fgr conformation, with the SH3 and SH2 domains engaging the SH2-kinase linker and C-terminal tail, respectively. In the Fgr:A-419259 complex, the activation loop of one monomer inserts into the active site of the other, providing a snapshot of trans-autophosphorylation. By contrast, TL02-59 binding induced SH2 domain displacement from the C-terminal tail and SH3 domain release from the linker. Solution studies using HDX MS were consistent with the crystal structures, with A-419259 reducing and TL02-59 enhancing solvent exposure of the SH3 domain. These structures demonstrate that allosteric connections between the kinase and regulatory domains of Src-family kinases are regulated by the ligand bound to the active site.
- Department of Microbiology and Molecular Genetics, University of Pittsburgh School of Medicine, Bridgeside Point II, Suite 523, 450 Technology Drive, Pittsburgh, PA 15219, USA.
Organizational Affiliation: